Genomic Diversity Decodes Evolutionary Mystery of "Living Fossil" Tree Ferns
Cyatheaceae tree ferns represent a typical ancient relict group within pteridophytes, drawing significant attention due to their archaic origins and distinctive tree-like forms. Their macroscopic morphology has remained highly conserved since the Jurassic period, earning them the reputation as "living fossils" from the age of dinosaurs. Despite their slow morphological evolution, this group exhibits remarkable species diversity and strong environmental adaptability, having successfully weathered multiple environmental upheavals over hundreds of millions of years. This longstanding puzzle in evolutionary biology remains unresolved: how could these lineages maintain such robust survival resilience while preserving stable morphological characteristics?
To address this, the Plant Evolution and Conservation research team at the South China Botanical Garden has constructed high-quality genome assemblies for different ecotypes of Cyatheaceae plants (including typical arborescent forms and non-arborescentforms lacking significant trunks). Through integrated comparative genomics and transcriptomics, they deeply investigated the underlying genome evolutionary mechanisms. The results indicate that, despite relatively slow overall molecular evolutionary rates, the entire tree fern lineage experienced only one shared whole-genome duplication (WGD) event approximately 154 million years ago. This event provided tree ferns with dual adaptive advantages—not only enabling ancestral tree ferns to adapt to late Jurassic environmental crises but also establishing the genetic foundation for subsequent species diversification.
The study also revealed that typical tree-forming species preferentially retained WGD-derived genes related to cell wall synthesis and lignification to reinforce support structures, while non-tree species retained more metabolism and defense-related genes to enhance survival resilience. In addition, continuous transposable element bursts have driven dynamic changes in genome architecture, including significant genome size variation and chromosomal rearrangements, serving as the core driver for local rapid evolution.
Furthermore, the innovation of arborescent phenotypes doesn't simply depend on WGD-retained gene dosage but relies more on complex transcriptional regulation. This mechanism likely facilitates tree fern adaptation to angiosperm-dominated understory environments by coordinating lignin synthesis with light perception. This research redefines the traditional concept of "evolutionary stasis" in living fossil plants as a state of genomic dynamic equilibrium. Genome plasticity and local innovation against a background of macroevolutionary conservatism represent the evolutionary key to tree ferns' successful persistence through hundreds of millions of years of geological changes.
Members of the research teamshared that these findings not only provide crucial genomic resources for conserving rare Cyatheaceae "living fossil" ferns but also establish a novel theoretical framework for understanding the long-term survival and evolution of ancient relict plants.
With support from the National Natural Science Foundation of China and other funding programs, Professors KANG Ming and WANG Jing from the South China Botanical Garden, Chinese Academy of Sciences, in collaboration with Professor Yves Van de Peer from Ghent University, Belgium, have deciphered the evolutionary mystery of Cyatheaceae "living fossil" tree ferns over hundreds of millions of years by analyzing their genomic diversity. This research was recently published in Molecular Biology and Evolution. Paper link: https://doi.org/10.1093/molbev/msaf247
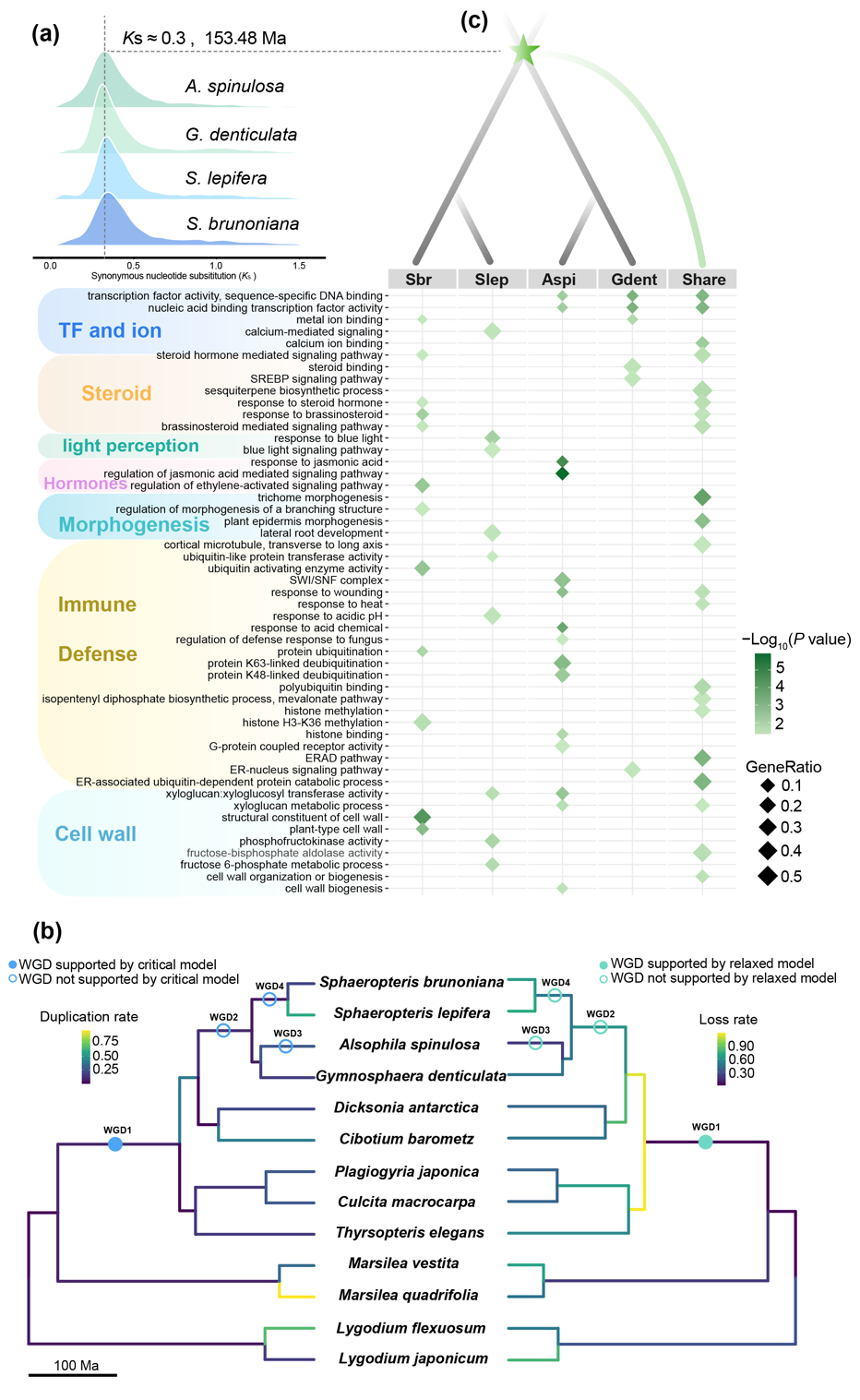
Figure 1. WGD events and subsequent gene retention across the four Cyatheaceae species. (Figure provided by the research team, same as below) (image by WANG et al)
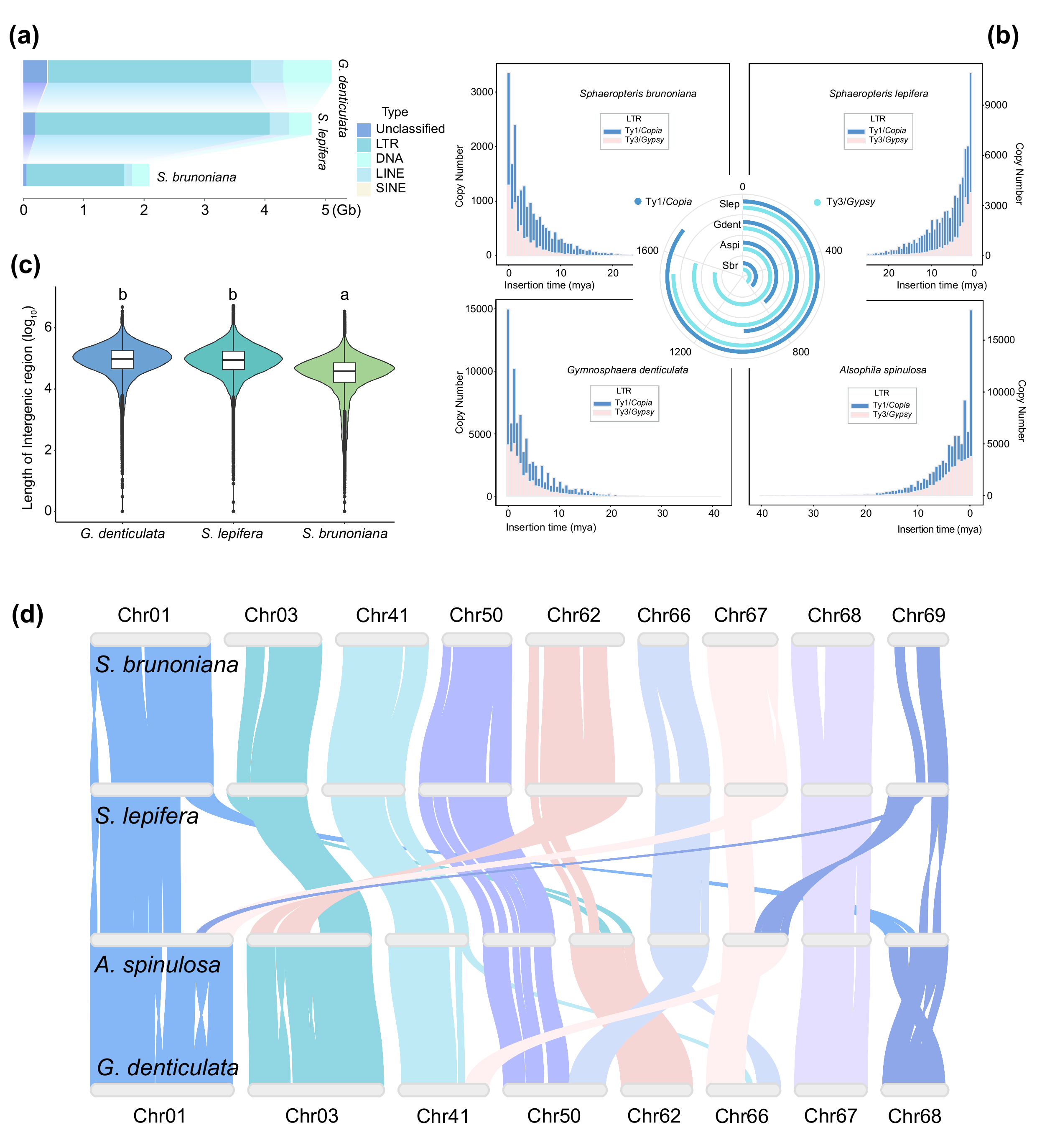
Figure 2. Genomic dynamism within Cyatheaceae. (image by WANG et al)
File Download: